Line of Demarcation Shown in Art Classical Lineshown in Art
1. Introduction
Over the past 4 decades, a grouping of perhabdoviruses (family Rhabdoviridae) have acquired losses of percid (perch and pike-perch) fry, juveniles and adults in European countries, generally on farms and in experimental facilities [ane,two,iii,4,five,6,7,viii,ix,10,eleven]. Ill fish usually brandish aberrant swimming behavior, such as swirling, alternating with lethargy. In some cases, hemorrhaging at the base of the fins can occur.
The Perhabdovirus genome is made upwards of a linear unmarried-stranded negative RNA of well-nigh eleven.5 kb encoding the 5 canonical rhabdovirus genes in the following society: nucleoprotein (N), polymerase-associated phosphoprotein (P), matrix protein (M), glycoprotein (Thou) and RNA-dependent RNA polymerase (L). For some viruses, additional small open reading frames (ORFs) are encoded, merely there are no experimental data illustrating their expression or role in the molecular biology of the virus. The genus Perhabdovirus includes 4 viral species recognized by the International Committee on Taxonomy of Viruses (ICTV): Perhabdovirus anguilla represented past viruses infecting eels only (i.east., EVEX), Perhabdovirus perca (formerly Perch perhabdovirus) and Perhabdovirus trutta (formerly Sea trout perhabdovirus), both represented past diverse viruses infecting percids every bit well as a few other fish species (i.e., Body of water trout rhabdovirus and virus R6146), and Perhabdovirus leman (including Leman rhabdovirus (LeRV)), described every bit virus xviii-193 [12]. Genomic data have demonstrated the existence of four main genogroups infecting percids. One genogroup is composed of all the viruses belonging to Perhabdovirus perca. A second cluster includes members of Perhabdovirus trutta. A third cluster is equanimous of a single virus (LeRV) representing Perhabdovirus leman. A fourth cluster includes a pair of viruses from France (18-203 and 18-206) that were initially thought to represent a new species considering the limited levels of nucleic acrid identities of the concatenated genes with the closest virus R6146 (fourscore%). Notwithstanding, because the new rules of demarcation of ICTV for perhabdoviruses, viruses xviii-203 and 18-206 should be classified every bit members of the Perhabdovirus trutta species since the levels of identity of their amino acid sequences with some members of this taxon are superior to 90%.
Considering this genetic diversity, iv sets of primers take previously been designed, each one able to dilate a specific portion of the N gene of all the members of a given genogroup [12]. Although the specificity of these assays is not high enough for diagnostic tests, because they occasionally produce non-specific products, they are useful as an N-based identification tool for any new isolate produced in cell culture or directly from infected fish.
Recently, accurate genotyping methods accept proved powerful for tracing some of the outbreaks of percid perhabdoviruses in Europe [7]. Nevertheless, molecular data are still scarce across the continent. Wild fishes, not only percids, stand for a reservoir for a number of perhabdoviruses, known and unknown, which are regularly introduced into farms or experimental facilities and are and then disseminated betwixt farms over Europe via international trade (eggs, fry, genitors). Advisable safety measures, such as egg disinfection, tin can help prevent the vertical transmission of these viruses, but they are not consistently employed by farmers. More generally, efficient diagnostic methods also lack for some of these viruses, and the few existing methods are insufficiently applied in European laboratories.
In 2019, an outbreak occurred in perch larvae on a farm in Western Europe, reaching a mortality charge per unit of almost thirty%. From sick fish, we isolated a perhabdovirus and obtained a near-complete sequence, which demonstrated the presence of a new genotype hitherto undescribed belonging to the Perhabdovirus perca species.
2. Results
2.1. Pathology
In December 2019, perch juveniles maintained in tanks in freshwater were affected by morbidities and mortalities. The mortality charge per unit was 0.3 to 1.v% daily, reaching well-nigh thirty% at the cease of the episode. A batch of fifty individuals was examined. All fishes had open gills and about 50% exhibited spinal distortion (Effigy ane). Aberrant swimming (swirling) was common. Several bacterial species were isolated, namely Aeromonas sp., Pseudomonas fluorescens and P. alcaligenes in the kidney, every bit well every bit Aeromonas sp., P. fluorescens and Providencia stuartii in the brain. No flavobacteriia were detected and no parasites were observed either in the gills, the skin, the fins or in the intestine.
2.2. Virus Detection
Seven days afterward inoculation of two prison cell lines with the supernatant of x fish basis together, a cytopathic effect typical of that induced by perhabdoviruses was observed. The jail cell layer was progressively destructed. After a 2nd passage of the supernatant of these, a strong ECP appeared only 24 h after inoculation. In another assay, after freezing at −80 °C for long-term storage, a 2d passage led to an ECP every bit soon as 4.five dpi with a complete destruction of the cells at 5.v dpi (Supplementary Figure S1). Because the host species affected by the disease and the symptoms (swirling), the presence of a perhabdovirus was suspected.
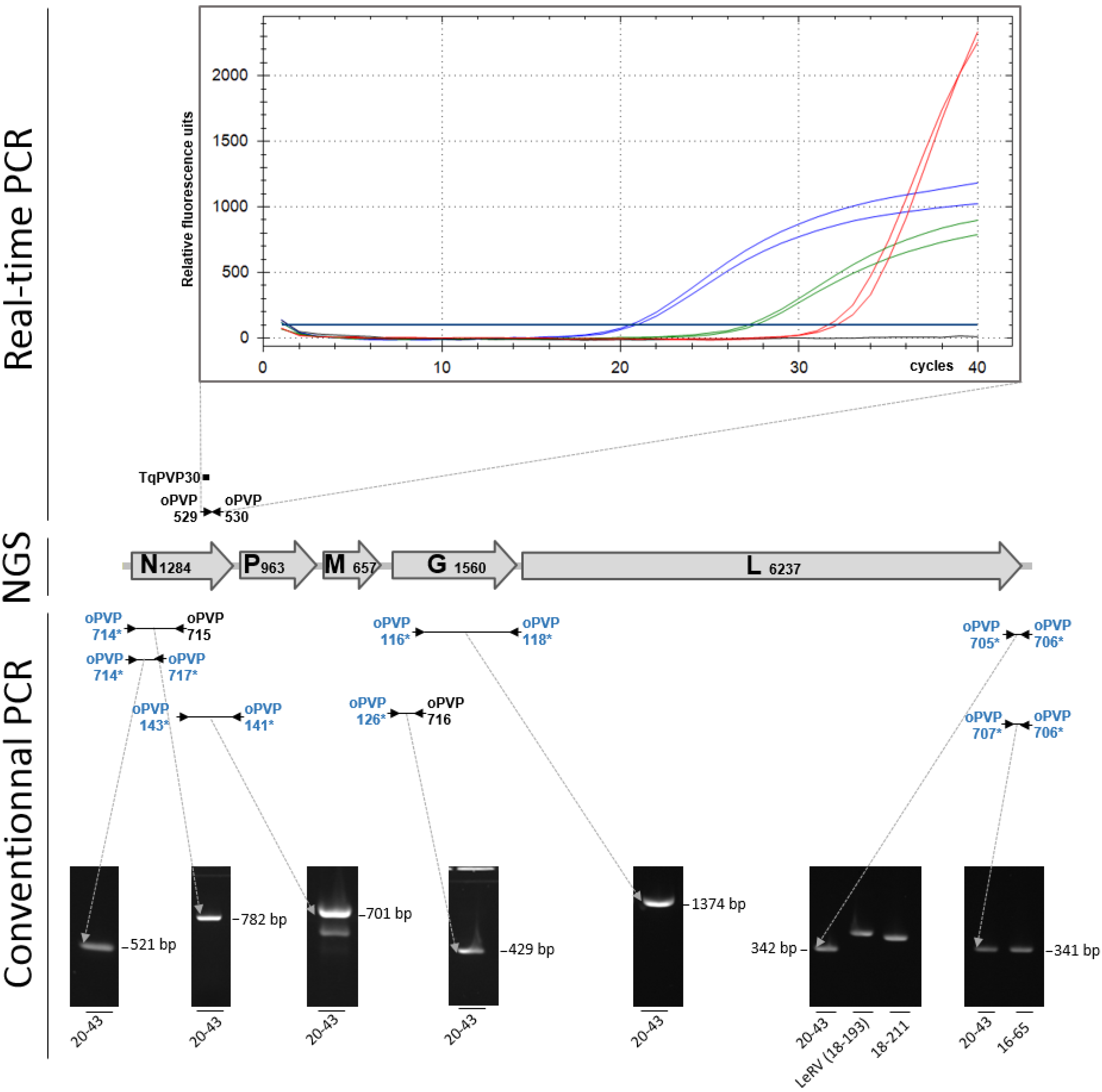
This viral isolate was named 20-43. Therefore, a series of specific RT-PCR assays was performed starting from nucleic acids extracted from the supernatants of cell cultures or directly from the footing organs. A real-time RT-PCR analysis targeting viruses of the Perhabdovirus perca species produced signals on both nucleic acid extracts, with Ct values betwixt 20.7 and 27.3 from cell civilization supernatants and organ extracts, respectively (Figure 2). All the same, the maximum levels of fluorescence of the amplification curves were lower than those obtained with a positive control, which exhibited a higher Ct value (31.9) and nigh two-fold the final fluorescence level compared with the isolate 20-43. This result suggested the presence of a perhabdovirus related to the tested genogroup, but its sequence likely had some mismatches that affected the point obtained with the real-fourth dimension RT-PCR primers and probe.
Information technology must be mentioned that, a few months subsequently the mortality episode affecting immature juveniles, three individual healthy fish, older than the ones tested above and maintained in a unlike site, were tested negative both past cell civilization and existent-fourth dimension RT-PCR assays (not shown). Furthermore, in 2021, 5 healthy private juveniles, from the progeny of genitors originating from the farm, were also tested negative by real-time RT-PCR. Therefore, healthy fish from two different generations were negative for this virus.
To genotype the virus, a range of conventional RT-PCR assays targeting the nucleoprotein genes of viruses from the iv known genogroups was performed as previously described. Surprisingly, no betoken was produced with whatever of the four RT-PCR assays, suggesting the virus 20-43 was relatively different from the known perhabdoviruses (not shown). Withal, an experimental generic cPCR targeting conserved portions of the iii′ cease of the percid perhabdoviruses and a fragment of the N ORF produced a clear signal of the expected size (Figure ii). A 2d generic assay (oPVP141/143) targeting the 3′ end of the North gene was also positive (Effigy 2). Similarly, two cPCR assays targeting a variable fragment of the 5′ end of the genome of percid perhabdoviruses each produced a point of size identical to the one expected for perch rhabdoviruses (Figure ii). All the same, for one particular assay (primer pair oPVP705/706), the size of the product was different from the size of the amplicons obtained with very distinct viral genotypes (Effigy 2). Finally, 2 generic RT-PCR assays targeting either a large portion of the Grand gene or the 3′ one-half of the N gene, besides produced bands at the expected sizes. All the sequences of these diverse genetic fragments confirmed the presence of a virus related to, though distinct from, perch rhabdovirus. These RT-PCR assays completed the missing portions of both Northward and G past combining strain-specific and generic primers (Figure ii). In summary, using primers amplifying different regions of perhabdoviruses, a total of seven amplicons were obtained whose sequences led to the complete N and G ORFs.
To sympathise why the genome of virus twenty-43 was non detected using cRT-PCR with any of the 4 sets of genogroup-specific primers, the primer sequences were aligned with the sequence of the N ORF newly obtained from virus 20-43. All primers exhibited several mismatches with their homologous sequence in virus twenty-43. For instance, there were a total of six mismatches between the N ORF of virus 20-43 and primers oPVP546 and oPVP547 targeting Perhabdovirus members. These numerous mismatches explained the absence of amplification signals for all the cRT-PCR primers. However, at that place were too some mismatches betwixt the N ORF and the oligonucleotides used for existent-time RT-PCR: two to iii for the primers, and ii for the TaqMan probe. It was, therefore, surprising that a bespeak was produced using real-fourth dimension RT-PCR, although the maximum fluorescence level of the amplification bend was lower than the one of a positive control perfectly adapted to the probe and primers.
ii.3. Genome Sequence
Past NGS sequencing the present viral isolate twenty-43, a sequence of xi,419 nt was obtained. This sequence did not contain the extremities of the genome but encompassed all the complete virus genes. As previously mentioned, a RT-cPCR assay targeting the 3′ stop of the genome led to the production of an amplicon, which once sequenced, added xv nt to the genomic sequence extending it to xi,434 nt. In comparison with the ends of the genomes of the near related viruses, it was speculated that 3 nt (possibly CGT) were missing at the 5′ finish of perhabdovirus 20-43, as well every bit 20 nt at the 3′ end, leading to an estimated size of 11,457 nt for virus 20-43. If correct, this size is like to those of the three sequenced members of the Perhabdovirus perca species and nearly 150 nt shorter than those of viruses R6146, xviii-193 and 18-203.
The genetic organization of virus 20-43 was typical of perhabdoviruses, with the five canonical genes Due north-Thousand-P-G-L (Figure 2). No additional small-scale ORFs (threshold 180 nt) were detected. The typical translation signal AACAG was present upstream of each ORF, and the termination transcription/polyadenylation (TTP) bespeak TATGA (7) was perfectly conserved downstream each ORF. Compared with related viruses from percids, the N ORF of virus xx-43 was slightly longer with a trinucleotide (CTG, Leucine) inserted at the terminate, just before the end codon. A trinucleotide insertion was also observed in the P ORF (963 nt) compared with the closest viruses members of Perhabdovirus perca xvi-65, 4890 and Perch rhabdovirus (960 nt). Even so, the P ORF was much shorter than its homologues from more afar viruses of the genus, for instance R6146 (993 nt). The M ORF had exactly the aforementioned size (657 nt) as viruses P8350 and 16-65, but it was slightly different from the respective ORF of other viruses such as LeRV (660 nt) and R6146 (642 nt). The length of the L ORF was strictly similar to that of the almost related viruses from percids.
2.four. Phylogenetic Studies
The consummate nucleotide sequence of the N ORF of virus 20-43 was aligned with a set of homologues belonging to the genus Perhabdovirus. It shared 69 to 81% of its nucleic acid identity with viruses from the genus Perhabdovirus, excluding EVEX which was more than distant (Effigy 3A). The most related viruses were those belonging to Perhabdovirus perca, for case French viruses M7173, 18-200 and xviii-195 every bit observed in the phylogenetic tree (Figure 3A). The N protein had a level of amino acid sequence identity of 89–92% with the N of viruses belonging to Perhabdovirus perca and less than 90% with the N of other perhabdoviruses (Effigy 3B). Because the Fifty ORF, the amino acid sequence identity of virus 20-43 varied between 80 and 91% identity with its homologues from other percid perhabdoviruses and identity just reached 72% with the L ORF of EVEX (Figure 3B).
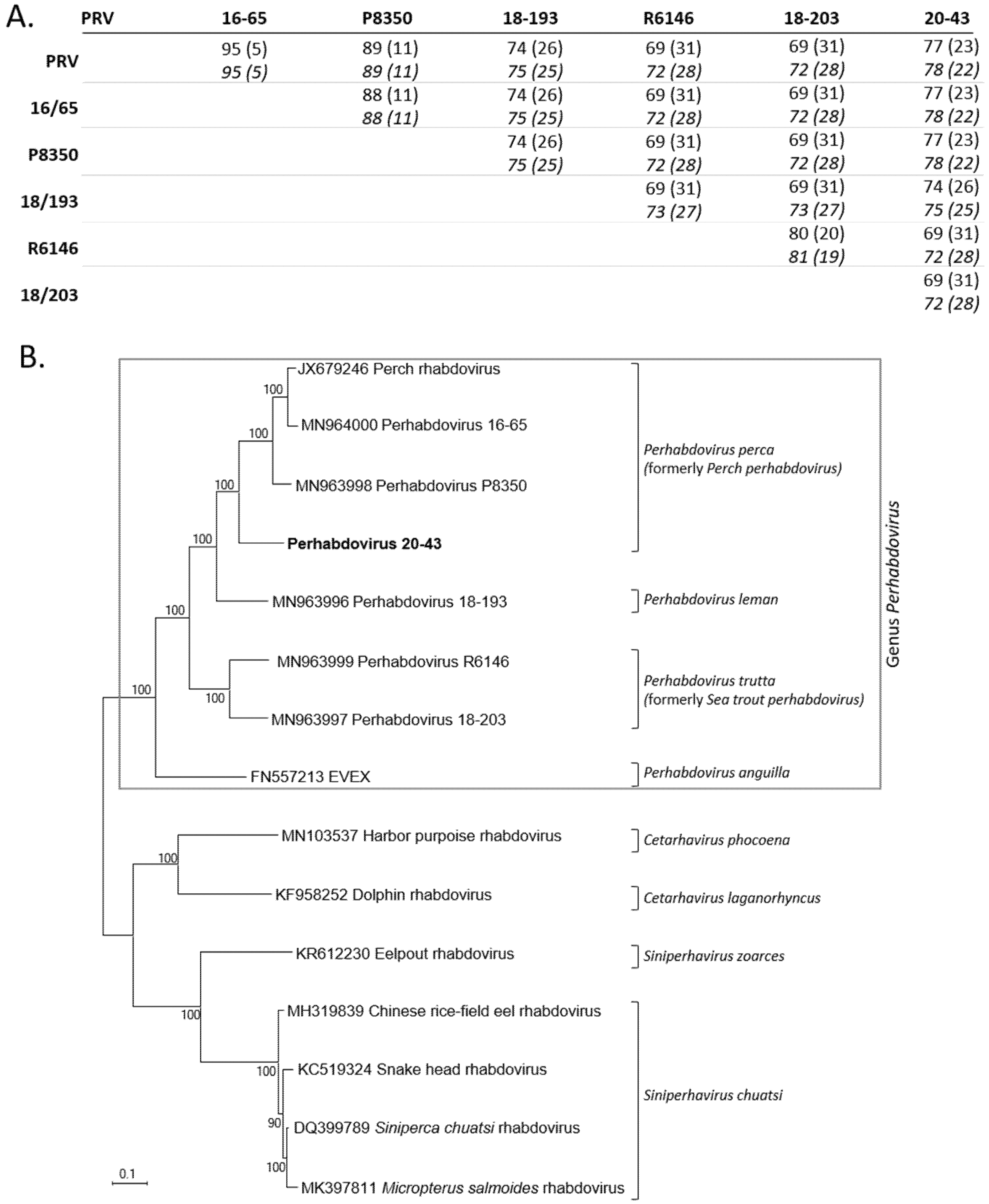
The concatenated nucleotide ORFs sequences were also aligned with the only six concatenated nucleotide ORFs sequences available from other related percid perhabdoviruses (Figure 4A). The identity levels of nucleic acids were 72–78% and 71–78%, respectively, with these six near related viruses. In parallel, we also aligned the complete nucleotide L ORF sequences alone and demonstrated the potent contribution of the 50 factor in the variability of the coding genome since the levels of identities of the Fifty and the concatenated ORFs were similar (Effigy 4A). The phylogenetic tree, based on the concatenated ORF sequences of these different viruses (Due north, P, Grand, G and L; 10107 positions compared) confirmed that virus 20-43 was included the genus Perhabdovirus together with other rhabdoviruses from percids (Figure 4B). More precisely, all these information point that the virus 20-43 belongs to the species Perhabdovirus perca (formerly Perch perhabdovirus). The 4 recognized species of the Perhabdovirus genus were within a clade distinct to another clade grouping rhabdoviruses from dolphin, tortoise or various fish species (Effigy 4B).
3. Give-and-take
A virus related to perhabdoviruses was isolated from perch juveniles affected by a mortality event on a European fish farm at the terminate of twelvemonth 2019. This virus was replicated in cell culture and was readily detected using a set of generic cRT-PCR assays and a species-specific real-time RT-PCR assay. Still, the virus was not detected using more specific assays targeting the four known genogroups of percid perhabdoviruses. This absence of detection was a posteriori explained past multiple mismatches between the viral sequence and the oligonucleotides. Its consummate sequence confirmed its human relationship to the other percid perhabdoviruses and its closest identities with viruses of the species Perhabdovirus perca, though there were numerous substitutions compared with the members of this species and with viruses from other species. Despite these nucleotide changes and regarding the threshold of poly peptide identity necessary to delineate 2 species (ninety%), we consider virus 20-43 as a member of Perhabdovirus perca, which is the most represented species in the outbreaks over the past decades. Withal, perhabdovirus 20-43 seems distinct plenty to represent a new viral lineage within the species.
The origin of perhabdovirus xx-43 is unknown, equally is its way of introduction into the farm. The farm is supplied with h2o from a jump, which cannot have been the source of the virus. Interestingly, several months before this episode, another mortality event affecting perch occurred. It happened just after connecting the water apportionment organization of tanks containing a few genitors captured in the wild to tanks containing fish from the farm. These latter fish were soon affected by morbidity and bloodshed. Unfortunately, no assay was performed on the dead fish from this first episode. Hence, there is a possibility that the wild fish were carriers of the virus and transmitted it horizontally to the perch produced in the farm via the water or fomites.
Regarding the bloodshed observed in December 2019 in this Western European fish farm, it affected fish merely afterwards their transfer to new tanks to start a new growing stage. Diverse bacteria were establish in the dead fish. Even so, they are generally considered as opportunistic and, therefore, likely not responsible of the mortality. A hypothesis is that virus 20-43 was at least partly responsible for the disease. The larvae were possibly already infected with the virus, albeit without whatsoever associated pathology. The manifestation of the infection may accept been triggered by either a modify in rearing conditions that subjected the fish to stress, therefore, making them more vulnerable, or, less likely, a sudden evolution of the virulence of this virus. From other episodes on other farms in Europe, it is known that the weaning of larvae and a modify in the feeding regime induces stress that can trigger viral replication. More often than not, the pathology of percid perhabdoviruses has been poorly studied and it is not clear why some members cause massive mortalities to larvae and others cause simply morbidity to juveniles and adults. More than research is needed to understand the virulence mechanisms of this viral genus.
A comprehensive overview of the multifariousness of the perhabdoviruses is needed to prevent their spread across Europe. The present genetic information likewise every bit distension methods can contribute to the rapid and authentic identification of whatsoever new isolate in order to compare it with known isolates, which is crucial for tracing outbreaks. Due to the full general lack of agile surveillance in Europe, it is likely that known, and yet unknown, genotypes and species will emerge in the side by side decade [12]. Furthermore, given the host-switching capacity of these viruses and their frequent geographic translocations, the emergence of any of these viruses will probable affect various hosts—non but those already recognized (percids and non-percids), but besides possibly new freshwater hosts that are increasingly farmed worldwide, such as tilapias.
4. Materials and Methods
iv.i. Virus Isolation
Samples were isolated in Dec 2019 from symptomatic Perca fluviatilis juveniles (size 3.3 cm; weight 0.four thou) maintained in freshwater at 16–18 °C. The supernatant of 10 fish footing together was used to inoculate two cultured jail cell lines, BF2 and RTG2 (ECACC, MERCK-SIGMA), equally already described [12]. Several months after this mortality episode, in June 2020, other fish from an older generation, salubrious and originating from the same farm, were tested for the presence of perhabdoviruses (Supplementary Figure S2). This batch was used every bit genitors to provide a new generation of juveniles. 5 of these juveniles, all healthy, were also submitted to a specific diagnostic test in September 2021.
4.2. Nucleic Acid Extraction, RT-PCR Detection and Sanger Sequencing
Full nucleic acids were extracted using a NucleoSpin Virus kit (Macherey-Nagel, Düren, Germany) with small-scale modifications every bit previously described [12]. The presence of a perhabdovirus was initially tested using a newly designed generic real-fourth dimension RT-PCR assay detecting various viruses of the Perhabdovirus perca species (Effigy 1). The pair of primers oPVP529 (GTGCAGGAARTCACCATACTCATC), oPVP530 (CAGCAGAGCACAGGTCATTTG) and a TaqMan probe, TqPVP30 (FAM-TCTGCCAATCCTGGAGTTCACTGCTG-BHQ1), were used at 0.4 µM in a final reaction book of 25 µL containing v µL of total nucleic acids. A SuperScript Three One-Step qRT-PCR kit (Invitrogen) was used with the post-obit thermo-cycling: a reverse-transcription stride of 30 min at 50 °C, followed by xv min at 95 °C and xl cycles of 15 sec at 94 °C and 60 sec at threescore °C. A viral isolate (16-121) highly related to Perch rhabdovirus was used equally a positive control [7].
Further molecular tests were performed using a range of RT-conventional PCRs (RT-cPCRs) combining generic and strain-specific primers (Figure 1 and Supplementary Table S1). For all the distension reactions, a SuperScript Three One-Step with Platinum Taq High-Allegiance kit (Invitrogen) was used according to the manufacturer's protocol by adapting the polymerization time to the length of the expected product and the melting temperature of the primers. Two overlapping fragments of the G gene were amplified as previously described [2]. Similarly, the complete North ORF was obtained by producing unlike amplicons, generic or strain-specific [2]. The 3′ finish of the genome was amplified using one primer (oPVP714) targeting a region that is highly conserved amid all the percid perhabdoviruses and another primer (oPVP717) targeting a moderately conserved region of N. The produced amplicon of 521 bp contained a part of the N ORF and a major part of the non-translated sequence upstream. The 5′ terminate of the viral genome was amplified using the two pairs of primers already published: oPVP705/oPVP706 and oPVP706/oPVP707 [12].
Later RT-PCR, 10 μl of these reactions was loaded on a two% agarose E-Gel (Invitrogen) and migrated 15 min earlier observation under UV light. Before sequencing, PCR products were purified using a NucleoSpin PCR clean-up kit (Macherey-Nagel) and subsequently, TA-cloned in a PCR4-TOPO vector (Invitrogen). For each amplicon, three clones were sequenced in both orientations with universal primers using the Sanger method and a 3130 Genetic Analyzer (Applied Biosystems).
four.3. Next-Generation Sequencing
To obtain the well-nigh-complete genome of the virus, next-generation sequencing (NGS) was performed using Illumina sequencing technology as previously described [12,13]. For the very few regions with low covering or uncertainties, Sanger sequencing was performed later on specific amplifications by RT-PCR. The complete viral genome sequence is available in GenBank (MW685822).
4.4. Phylogenetic Analysis
Deoxyribonucleic acid sequences obtained from Sanger sequencing were assembled and the consensus sequence edited using VectorNTI11.5 (Invitrogen). The identities levels were obtained past adjustment the sequences using the 'marshal' function of VectorNTI and the identity matrix was constructed using the RStudio software. For the phylogenetic analyses, the sequences were aligned with the ClustalW office of MEGA7 or MEGA10 [14,fifteen]. Maximum-likelihood (ML) analyses were performed using a transition-to-transversion ratio of 2.0 with empirical base frequencies and the HKY85 commutation model implemented in the MEGA program [14]. Bootstrap analyses were performed using 1000 replications. Values greater than 90% were considered strong evidence for robust phylogenetic groupings.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/x.3390/pathogens10101256/s1, Figure S1: Cytopathic event associated with the perhabdovirus twenty-43. Cells are BF2 observed several days afterward infection (pi) (A) 4.v days pi. (B) five.5 days pi. (C) negative command 4.v days pi. Figure S2: Sampling of the fish. Table S1: Primers and probe used in this study.
Author Contributions
Conceptualization, L.B., F.P., A.50.; data curation, L.B., L.P., L.D.; formal analysis, L.B., L.P., L.D.; funding conquering, Fifty.B., F.P.; investigation, Fifty.P., C.F., F.P., 50.D., L.B.; project administration, L.B., F.P.; supervision, L.B., F.P., L.D.; writing—original draft, L.B.; writing—review and editing, F.P., Fifty.D., L.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Maritime and Fisheries Fund (projection PERCI-HATCH). The funder had no function in the design of the study; in the collection, assay, or interpretation of information; in the writing of the manuscript or in the decision to publish the results.
Institutional Review Lath Statement
Not applicative.
Informed Consent Statement
Not applicative.
Data Availability Statement
The new genetic data are available in GenBank (MW685822).
Acknowledgments
We acknowledge the sequencing facilities at the ANSES Ploufragan-Plouzané-Niort laboratory and the 'Plate-forme de microbiologie mutalisée (P2M)' at the Institut Pasteur (Paris) for technical assist with NGS sequencing. We thank Stéphanie Bougeard for her invaluable help in preparing Figure 3. Peter Walker and Gael Kurath are acknowledged for their invaluable help in proposing a new classification of the perhabdoviruses. Yannick Ledoré is acknowledged for providing healthy fish every bit controls. Carolyn Engel-Gautier is acknowledged for correcting the English of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gadd, T.; Viljamaa-Dirks, S.; Holopainen, R.; Koski, P.; Jakava-Viljanen, 1000. Characterization of perch rhabdovirus (PRV) in farmed grayling Thymallus thymallus. Dis. Aquat. Organ. 2013, 106, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Talbi, C.; Cabon, J.; Baud, K.; Bourjaily, M.; de Boisseson, C.; Castric, J.; Bigarré, 50. Genetic diverseness of perch rhabdoviruses isolates based on the nucleoprotein and glycoprotein genes. Arch. Virol. 2011, 156, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- Betts, A.M.; Rock, D.Thou.; Way, K.; Torhy, C.; Chilmonczyk, Southward.; Benmansour, A.; de Kinkelin, P. Emerging vesiculo-type virus infections of freshwater fishes in Europe. Dis. Aquat. Organ. 2003, 57, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Wahli, T.; Bellec, 50.; von Siebenthal, B.; Cabon, J.; Schmidt-Posthaus, H.; Morin, T. Offset isolation of a rhabdovirus from perch Perca fluviatilis in Switzerland. Dis. Aquat. Organ. 2015, 116, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Ruane, Due north.M.; Rodger, H.D.; McCarthy, L.J.; Swords, D.; Dodge, Chiliad.; Kerr, R.C.; Henshilwood, G.; Stone, D.M. Genetic diversity and associated pathology of rhabdovirus infections in farmed and wild perch Perca fluviatilis in Ireland. Dis. Aquat. Organ. 2014, 112, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Dannevig, H.; Olesen, N.J.; Jentoft, S.; Kvellestad, A.; Taksdal, T.; Hastein, T. The commencement isolation of a rhabdovirus from perch (Perca fluviatilis) in Norway. Balderdash. Eur. Assoc. Fish Pathol. 2001, 21, 186–194. [Google Scholar]
- Bigarré, L.; Plassiart, G.; de Boisseson, C.; Pallandre, L.; Pozet, F.; Ledore, Y.; Fontaine, P.; Lieffrig, F. Molecular investigations of outbreaks of Perch perhabdovirus infections in freeway-perch. Dis. Aquat. Organ. 2017, 127, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Nougayrède, P.; de Kinkelin, P.; Chilmonczyk, S.; Vuillaume, A. Isolation of a rhabdovirus from the expressway-perch Stizostedion lucioperca (L.1758). Bull. Eur. Assoc. Fish Pathol. 1992, 12, v–seven. [Google Scholar]
- Dorson, Grand.; Torchy, C.; Chilmonczyk, Southward.; de Kinkelin, P. A rhabdovirus pathogenic for perch, Perca fluviatilis L.: Isolation and preliminary study. J. Fish Dis. 1984, 7, 241–245. [Google Scholar] [CrossRef]
- Jorgensen, P.E.V.; Olesen, North.J.; Ahne, West.; Wahli, T.; Meier, West. Isolation of a previously undescribed rhabdovirus from pike Esox lucius. Dis. Aquat. Organ. 1993, 16, 171–179. [Google Scholar] [CrossRef]
- Caruso, C.; Gustinelli, A.; Pastorino, P.; Acutis, P.L.; Prato, R.; Masoero, L.; Peletto, Southward.; Fioravanti, Yard.50.; Prearo, M. Mortality outbreak by perch rhabdovirus in European perch (Perca fluviatilis) farmed in Italia: Clinical presentation and phylogenetic assay. J. Fish Dis. 2019, 42, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Pallandre, Fifty.; Luo, D.; Feuvrier, C.; Lieffrig, F.; Pozet, F.; Dacheux, L.; Bigarré, L. Revisiting the classification of percid perhabdoviruses using new full-length genomes. Viruses 2020, 12, 649. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.S.; Li, B.; Shen, X.R.; Jiang, R.D.; Zhu, Y.; Wu, J.; Fan, Y.; Bourhy, H.; Hu, B.; Ge, X.Y.; et al. Characterization of Novel Rhabdoviruses in Chinese Bats. Viruses 2021, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, Chiliad. MEGA7: Molecular Evolutionary Genetics Analysis Version seven.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kumar, South.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Calculating Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
Figure 1. Perca fluviatilis juveniles affected by a mortality episode in a fish farm from Western Europe in 2019. The arrow shows the open up gills observed in affected animals.
Figure one. Perca fluviatilis juveniles afflicted by a mortality episode in a fish farm from Western Europe in 2019. The arrow shows the open gills observed in afflicted animals.

Figure 2. Molecular detection and genetic characterization of virus 20-43. The almost-complete sequence of the viral genome was obtained by combining adjacent-generation sequencing (NGS) and conventional PCR (cPCR) (come across text). ORFs and their size (in nt) are indicated on the map. A real-time RT-PCR targeting viruses belonging to the Perhabdovirus perca species gave curves showing an unusual shape compared with controls (blue lines, curves obtained from cell cultures infected with xx-43; dark-green lines, curves obtained from infected fish organs; cherry lines, curves of a positive control represented past a strain of perch perhabdovirus); the three samples are in duplicate. Diverse cPCRs (night lines) were used to confirm the presence of a perhabdovirus in the inoculated prison cell culture. Primers are symbolized by arrowheads. Generic primers (in bluish) targeting various percid perhabdoviruses are labelled with asterisks; specific primers designed from the sequence of virus xx-43 are in black. 3 related perhabdoviruses were used every bit controls for the amplifications of a region in the 5′ end of the genome [12]. Note the different sizes of the amplicons for viruses 18-193 and 18-211 compared to virus xx-43.
Figure 2. Molecular detection and genetic characterization of virus 20-43. The near-complete sequence of the viral genome was obtained by combining next-generation sequencing (NGS) and conventional PCR (cPCR) (see text). ORFs and their size (in nt) are indicated on the map. A real-time RT-PCR targeting viruses belonging to the Perhabdovirus perca species gave curves showing an unusual shape compared with controls (blue lines, curves obtained from prison cell cultures infected with xx-43; green lines, curves obtained from infected fish organs; cherry lines, curves of a positive command represented by a strain of perch perhabdovirus); the iii samples are in duplicate. Various cPCRs (dark lines) were used to confirm the presence of a perhabdovirus in the inoculated cell culture. Primers are symbolized by arrowheads. Generic primers (in blue) targeting diverse percid perhabdoviruses are labelled with asterisks; specific primers designed from the sequence of virus 20-43 are in black. Iii related perhabdoviruses were used equally controls for the amplifications of a region in the five′ terminate of the genome [12]. Note the different sizes of the amplicons for viruses eighteen-193 and 18-211 compared to virus 20-43.
Effigy 3. Comparison of selected ORFs of virus 20-43 with homologous sequences of perhabdoviruses. A. The maximum-likelihood phylogenetic tree compares the nucleic acid sequences (MEGA7, 1284 positions compared) of the N ORF from a range of viruses of dissimilar origins (countries indicated with their ii-letter code) belonging to the genus Perhabdovirus. EVEX was used as an outgroup. Identities are color-coded in the oestrus map matrix. B. Amino-acid sequence identities (amino-acid sequences) of the L (black) and Due north (bluish italics) ORFs of perhabdoviruses for which both of these sequences are available.
Figure three. Comparison of selected ORFs of virus 20-43 with homologous sequences of perhabdoviruses. A. The maximum-likelihood phylogenetic tree compares the nucleic acid sequences (MEGA7, 1284 positions compared) of the N ORF from a range of viruses of dissimilar origins (countries indicated with their two-letter code) belonging to the genus Perhabdovirus. EVEX was used as an outgroup. Identities are colour-coded in the heat map matrix. B. Amino-acid sequence identities (amino-acid sequences) of the L (black) and Due north (blue italics) ORFs of perhabdoviruses for which both of these sequences are available.
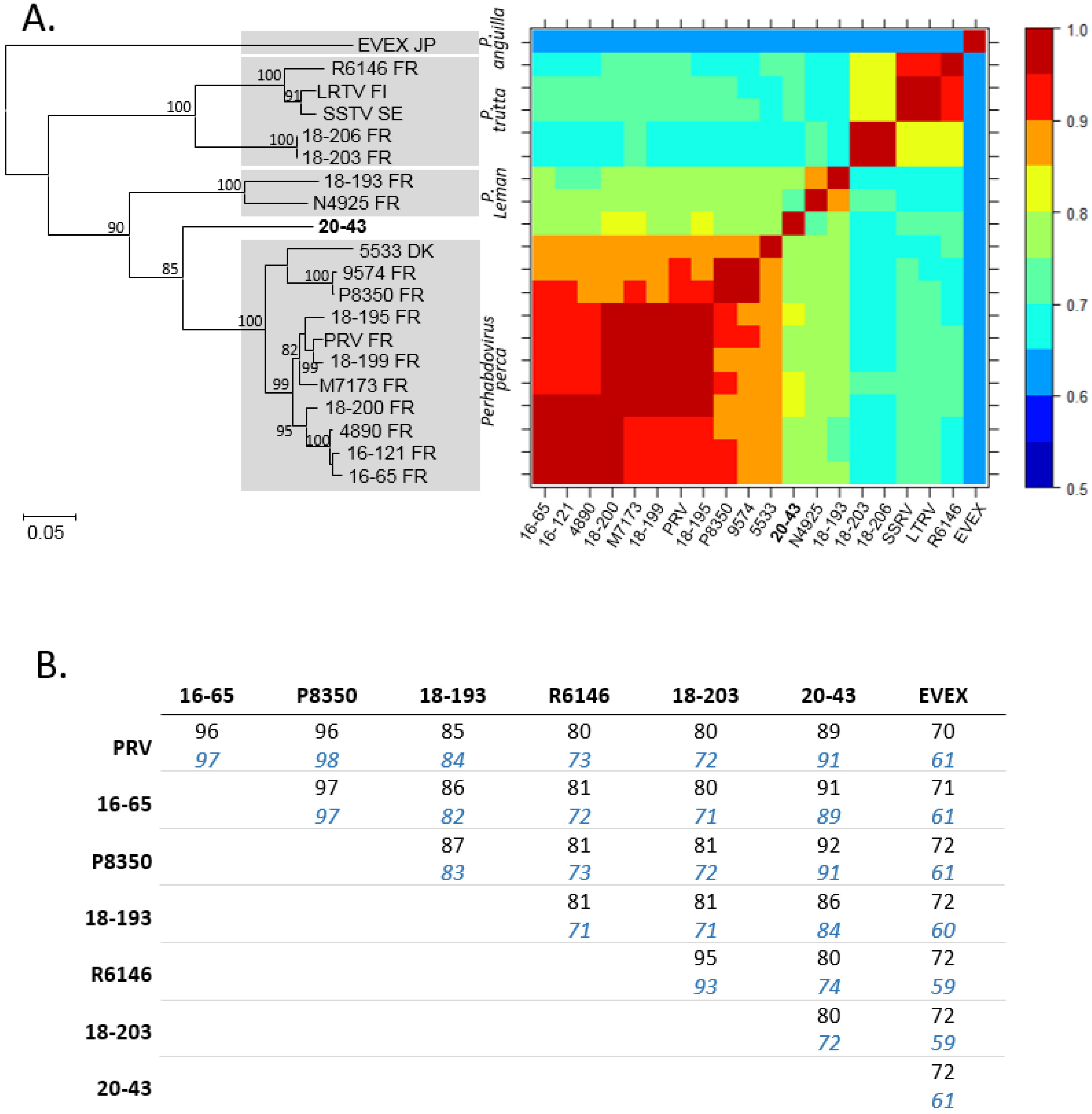
Figure 4. Comparisons and genetic relationships betwixt concatenated homologous ORFs of percid perhabdoviruses and other fish rhabdoviruses. A. Pairwise nucleic acid identities in % (divergence in parentheses) of the concatenated ORFs (starting time line) and the L ORF only (2d line, shown in italics). B. Maximum-likelihood phylogeny (1000 bootstraps with MEGA10 software) of concatenated ORF sequences (Due north, P, Grand, G and L) of virus 20-43 (in assuming) and homologous sequences of perhabdoviruses and other fish rhabdoviruses (10107 positions compared). The scale bar indicates genetic distance (substitutions/site).
Effigy 4. Comparisons and genetic relationships between concatenated homologous ORFs of percid perhabdoviruses and other fish rhabdoviruses. A. Pairwise nucleic acid identities in % (divergence in parentheses) of the concatenated ORFs (first line) and the L ORF only (2nd line, shown in italics). B. Maximum-likelihood phylogeny (1000 bootstraps with MEGA10 software) of concatenated ORF sequences (N, P, K, G and L) of virus twenty-43 (in bold) and homologous sequences of perhabdoviruses and other fish rhabdoviruses (10107 positions compared). The scale bar indicates genetic distance (substitutions/site).
| Publisher's Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This commodity is an open access commodity distributed under the terms and conditions of the Creative Commons Attribution (CC Past) license (https://creativecommons.org/licenses/by/iv.0/).
Source: https://www.mdpi.com/2076-0817/10/10/1256/htm
Belum ada Komentar untuk "Line of Demarcation Shown in Art Classical Lineshown in Art"
Posting Komentar